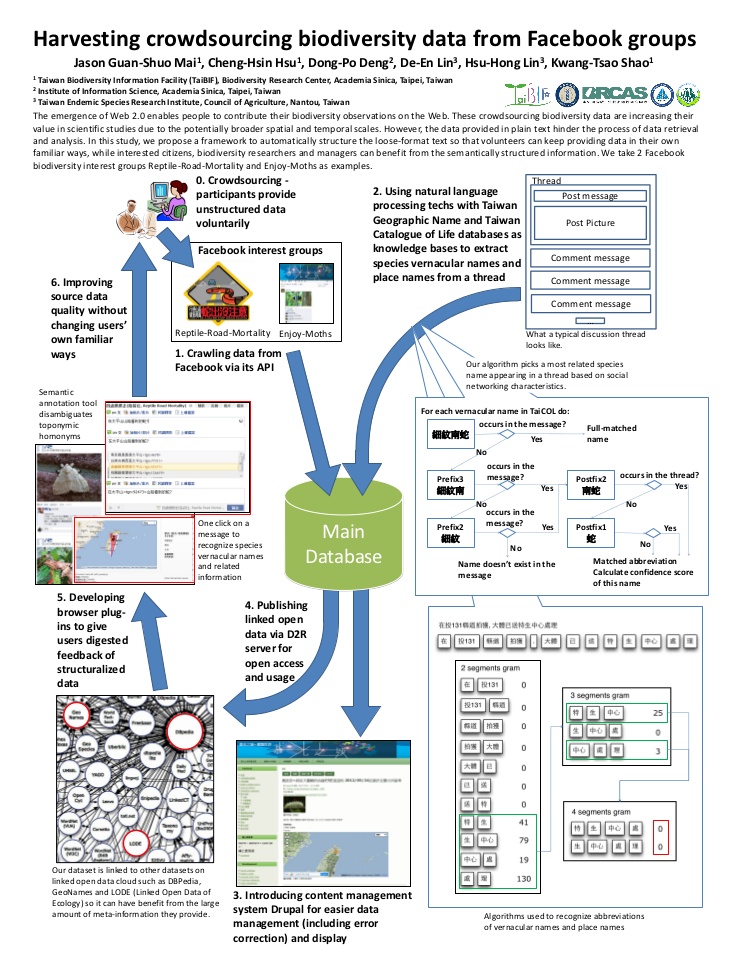
Volunteer participation in scientific studies is increasing their value in the past decade due to the emergence of Web 2.0 technologies. Volunteer naturalists produce crowdsourced information about biodiversity through participative online activities. “Reptile‐Road‐Mortality” and “Enjoy‐Moths” are both interest groups on Facebook as well as citizen science projects. The former provides a platform for reporting road‐kill events, while the latter is for moth observations in Taiwan. People “post” photographs with meta‐information and experienced participants help identifying species in the “comments”. However, most crowdsourcing information is unstructured. The interface is easy for participating but hard for organizing data. Here we propose an approach to make crowdsourced biodiversity data more suitable for scientific use. Three technological components are included: natural language processing (NLP), semantic web, and content management system (CMS). Natural language processing is applied to extract species names, locations, and times, in the descriptions of photographs and comments. Extracted information would be encoded to a resource description framework (RDF) and published as linked data for open access. Meanwhile, data are imported into CMS for efficient managing and displaying. By applying this approach, scientists can easily analyze the patterns of the occurrence of road‐killedspecies and moths while keeping the way in which volunteers contribute unchanged.
downlowad the all abstracts of The 2nd Asian Regional Conference of Society for Conservation Biology